RESEARCH
Computational tool for designing internal probes with matching primer sets for real-time PCR experiments and melting curve analysis of genetic variations
Edesign, an efficient computational tool for designing internal probes with matching primer sets for real-time PCR experiments and melting curve analysis of genetic variations, has been developed by researchers at RIKEN and the Japanese firm K.K.DNAFORM. This tool will enable the development of new, reliable assays for DNA-based genetic testing, and help to bring the benefits of genome-wide sequence analysis to patients in the clinic.
Yasumasa Kimura, Takahiro Soma, and colleagues from the RIKEN Center for Life Science Technologies, and the RIKEN Preventive Medicine & Diagnosis Innovation Program, and K.K.DNAFORM report today on a new internal probe and PCR primer set design tool for real-time PCR and melting curve experiments in the journal PLOS ONE.
PCR, for Polymerase Chain Reaction, is a simple and inexpensive DNA amplification technique, widely used to analyze DNA and RNA in medical diagnostics, life science laboratories and industrial applications. During PCR, small amounts of target DNA molecules are copied and rapidly amplified, thus enabling researchers to analyze or clone DNA. The PCR amplification products can be detected and further characterized by addition of internal probes to the reaction mixture. These internal probes hybridize to specific sequences within the amplified DNA and have defined melting temperatures for the separation of the internal probe from its binding sequence in the target DNA molecule. Internal probes are used to monitor amplification reactions (“real-time PCR”), and to distinguish between different amplification products in the same reaction mixture (“multiplex PCR”). Even single-base pair sequence variations between the internal probe and a target DNA molecule can be reliably detected by melting curve analysis. Therefore real-time PCR and melting curve analysis are applied in the clinical routine to diagnose diseases, and are the basis to many molecular diagnostic tests in the market.
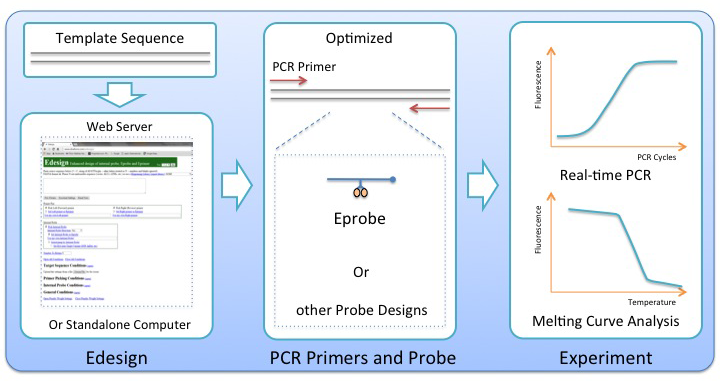
While real-time PCR and melting curve analysis are among the most powerful tools used in the laboratory, the quality of any such assay is largely dependent on careful selection of the amplification primers and a matching internal probe. Because of the high complexity of all parameters to be considered during the assay development, primer and probe designs are commonly aided by computational tools made available by the research community or offered by commercial vendors. While many PCR primer design tools are available in the public domain, there is an unmet need for a better platform to do integrated designs of primer sets together with matching internal probes and to support new technologies like high-resolution melting curve analysis, and the use of new generations of internal probes like Eprobes during PCR. Eprobes have been developed at RIKEN and K.K.DNAFORM as hybridization probes for PCR experiments and the detection of genetic variations. They use a new chemistry for complete signal suppression in the single-stranded form, and enhanced DNA-binding affinity caused by intercalation of the dye moieties into double-stranded DNA after hybridization to its target sequence. To increase the DNA-binding affinity of an internal probe is an essential advantage for the selective discrimination of single-base pair variations during melting curve analysis.
Based on the established primer design software Primer3, we added in Edesign specific functions for the design of a matching internal probe, which allow for binding of the internal probe to either strand in the target DNA molecule, and to consider possible interference between the internal probe and the amplification primers. The user has multiple options in the interface to adjust the settings for example for the melting temperature of the internal probe, and to define the regions where the internal probe and primers should bind within the target sequence. Edesign can provide designs for any kind of DNA-based internal probe, like for instance the commonly used TaqMan probes, but additional features were developed to extend the program with the physical parameters of internal probes using a different chemistry altering the DNA-binding affinity or other properties of an internal probe. These features have been used to add to Edesign the possibility to design also thiazole orange-labeled Eprobes for use as internal probes. During the studies, physical properties of thiazole orange-labeled Eprobes were analyzed in detail to obtain the necessary parameters for updating the reference tables in Edesign. The integration of thiazole orange-labeled Eprobes into Edesign serves as a reference on how any other type of probe chemistry could be added to Edesign making it a universal tool for integrated designs of internal probes with matching primer sets. Particular care has been taken to include into Edesign features for flexible but effective design of internal probes intended for genotyping experiments.
RIKEN made the source code for Edesign available for download at https://github.com/yasumasak/edesign/ under the terms of a GNU General Public License version 2.0 (GPLv2); K.K.DNAFORM maintains an open Edesign web interface for unlimited public use at http://www.dnaform.com/edesign2/.
“Although, internal probe usage in real-time PCR has been around for a long time, no public software really has integrated all the different aspects of internal probe design such as length, presence of mismatches, complementarity to primers, to mention a few, and therefore it could have been a manual painstaking exercise to design primer-probe pairs. Thus, Edesign is a real advance in the field to support the research community. In particular, parameters measured experimentally were fed into the algorithms to substantially improve the design of primer-Eprobe sets.” explains Yasumasa Kimura, one of the first authors of the publication.
“Eprobes are very flexible and can be applied both for real-time PCR and melting curve analyses, thus offering advantages for several medical needs becoming possible by gene-based diagnostic assays. The computational “Edesign” tool for automatic and precise design of Eprobes will contribute to rapidly developing new Eprobe-based diagnosis kit and hence help expanding the number of available gene diagnosis tests for clinical use.” explains Kengo Usui, the leader of the Genetic Diagnosis Technology Unit at RIKEN Center for Life Science Technologies.
“The Edesign software is free to download and therefore we hope that it will be widely used by the community.”, comments Matthias Harbers, Visiting Scientist to the Division of Genomic Technologies at the RIKEN Center for Life Science Technologies and one of the corresponding authors. “Besides using it for working with Eprobes and other DNA-based internal probes, we further provide for Edesign a tool to update the software with the thermodynamic data for designing other types of internal probes thus making Edesign an universal application for primer and probe designs.”
Reference
Contact
Kengo Usui, Unit Leader
Genetic Diagnosis Technology Unit
Omics Application Technology Group
Division of Genomic Technologies
RIKEN Center for Life Science Technologies