Team Leader

Establishment of a structural analysis technology platform that contributes to "life innovation" such as drug development and medical treatment.
* Due to the reorganization starting as new centers in April 2018, this laboratory is now belong to the Center for Biosystems Dynamics Research. As for the latest information, please see the following URL below.
> The webpage of Laboratory for Protein Functional and Structural Biology, Center for Biosystems Dynamics Research

Team Leader
Mikako Shirouzu
#W209 West Research Building, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama City, Kanagawa, 230-0045, Japan
Tel: +81-(0)45-503-9202
![]()
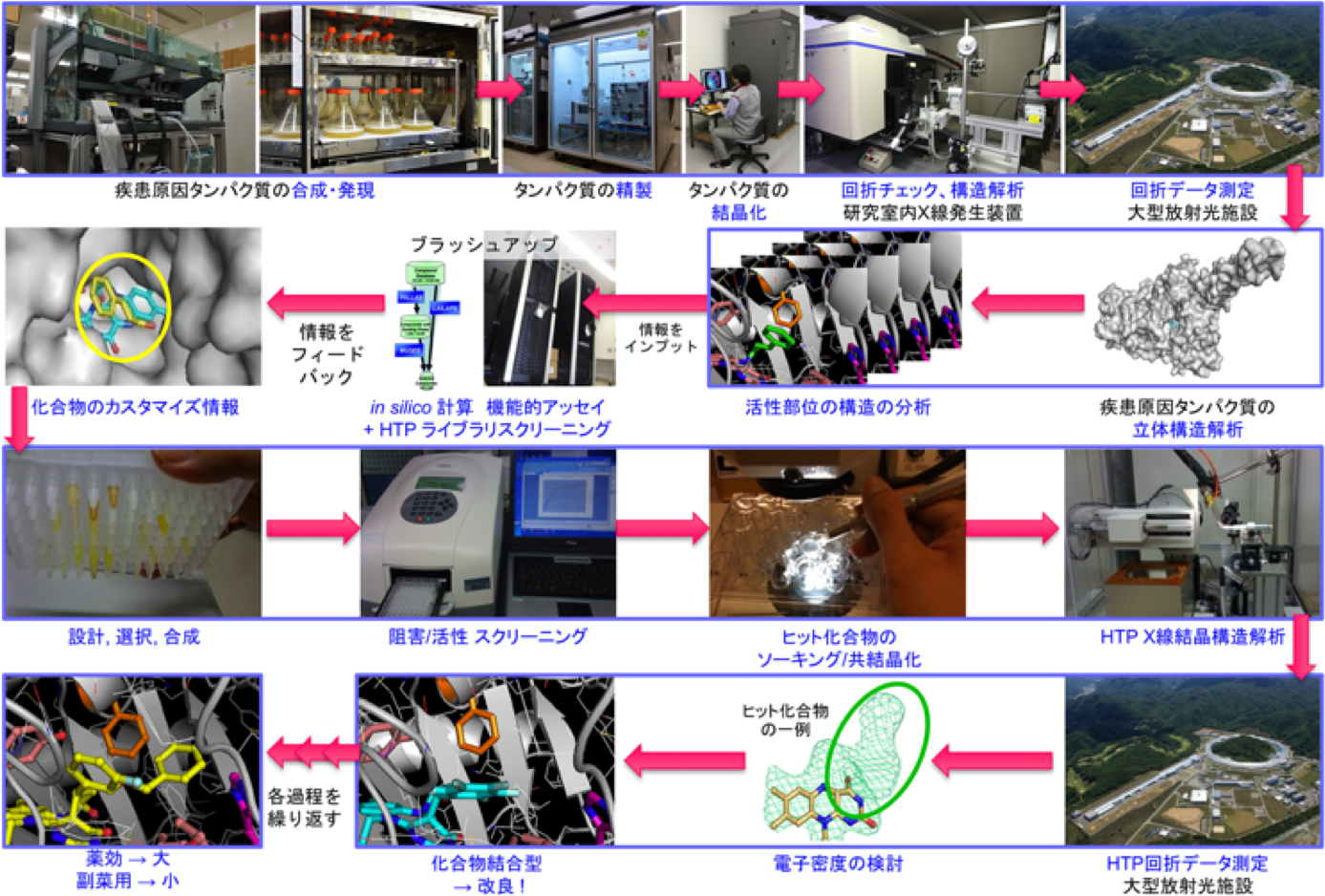
We plan to establish a structural analysis technology platform to contribute to "life innovation" such as drug discovery and medical treatment as well as develop methods for the sample preparation and crystallization of challenging proteins including membrane proteins that have been difficult in its analysis. The high-resolution structural information of proteins related to diseases shall increasingly become important for drug development leading to future individualized medicine such as a molecular target drug. Especially, we will perform structural analysis mainly for proteins related to immune system diseases and kinases associated with cancer/leukemia. The 3D-structural information of these target proteins will be used for in silico screening of chemical compounds. Further analysis of the complex of the target protein with candidate compounds will provide valuable information for drug discovery.
CLST was reorganized into three centers according to the RIKEN 4th Medium-Term Plan from April 1, 2018. For the latest information of Protein Functional and Structural Biology Team, please visit the following websites.
> The webpage of Laboratory for Protein Functional and Structural Biology, Center for Biosystems Dynamics Research [http://www.bdr.riken.jp/en/research/labs/shirouzu-m-protein/index.html]]
> If you continue to browse this site, click here.