Team Leader

* Due to the reorganization starting as new centers in April 2018, this laboratory is now belong to the Center for Biosystems Dynamics Research. As for the latest information, please see the following URL below.
> The webpage of Laboratory for Structure-Based Molecular Design, Center for Biosystems Dynamics Research

Team Leader
Teruki Honma
1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045 JAPAN
Tel: 045-503-9433
![]()
We plan to develop methods for designing novel small molecules, nucleic acids, and proteins, in order to help to accelerate drug development and the elucidation of living systems, and also develop an FBDD (Fragment based Drug Discovery) method for targeting biomolecules currently difficult to make susceptible of binding to small molecules.
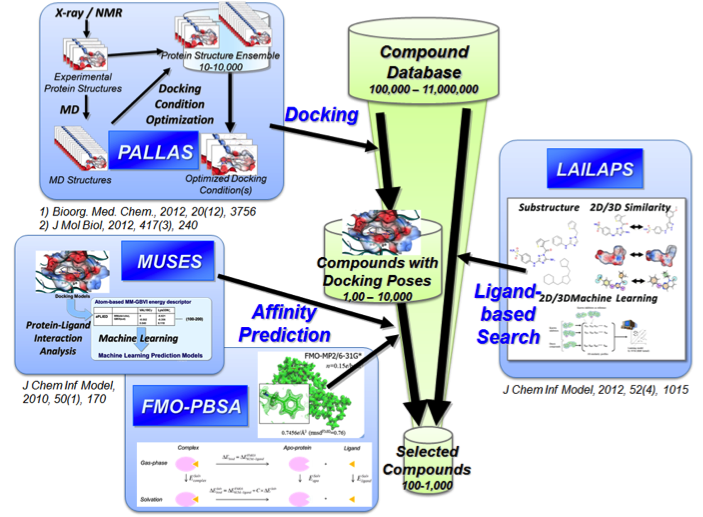

CLST was reorganized into three centers according to the RIKEN 4th Medium-Term Plan from April 1, 2018. For the latest information of Structure-based Molecular Design Team, please visit the following websites.
> The webpage of Laboratory for Structure-Based Molecular Design, Center for Biosystems Dynamics Research [http://www.bdr.riken.jp/en/research/labs/honma-t-labo/index.html]
> If you continue to browse this site, click here.